Welcome to the AGRIS tutorial on the ATTFDB
The following tutorial was
developed to help guide a new user through conducting a search through the ATTFDB and provides information on
using the website as a whole.
AGRIS
Homepage-
The header contains links to
“HOME” the homepage, the “ATCISDB” a cis-regulatory database consisting of the
5' regulatory sequences of all 25,080 annotated genes with a description of the corresponding
cis-regulatory elements. The “ATTFDB” a database of information on almost 1,550
transcription factors
(TF) arranged into 39 families. “NSF2010” will provide information about the NSF
information of Arabidopsis 2010 project, “DOWNLOADS” will provide information
about the data download information for AtcisDB and AtTFDB, “PEOPLE” will provide
information about the individuals that have currently contributed to the
website, “LINKS” contains links to additional Arabidopsis resources, “NEWS”
will provide information about recent updates and events. In addition, there
are links to various departments at The Ohio State University that have
contributed to the project.
Search the ATTFDB
Click on the ATTFDB link

ATTFDB Homepage-
On the ATTFDB homepage, a search can be conducted in two
ways – through a specific search or through browsing by family.
Specific Search: using one of the following
·
Locus ID: a
unique locus name that corresponds to a transcribed region in the Arabidopsis
genome. The names were designated as
part of the genome sequence annotation and are associated with genes, genetic
markers, polymorphic states, or map features.
The format of the LocusID is, AtXgYYYYY. The “At” refers to Arabidopsis thaliana, the “X” number corresponds
to the chromosome number, the “g” refers to gene and the 5 following “Y”s
correspond to the five-digit code, numbered from top to bottom of the
chromosome.
·
Part of Description: using part of the description or gene name
And then click “Search”
Browse
Families: select on of the 39 families listed to see all transcription
factors currently identified.

Search Results Page-
Search
Results are listed as a table.

Family
Info:
Name of the family – “MADS, MYB”
Description – Description of the family
Reference –
Reference information for the specific family
ClustAlW Alignment – ClustAlW alignment for
protein sequences in the family
Hidden Markov Model – Hidden Markov Model for
the entire family
Nucleotide Sequences – Nucleotide sequences
for the genes in the family
Protein Sequences – Protein sequences for the
genes in the family
Members identified for a family
Locus Id – Locus Id of the gene
Gene Name – Name of the gene
Links – Links to T (TIGR), M (MIPS), S (SALK),
T (TAIR) websites for the specific gene
Direct Targets – Confirmed
/ Unconfirmed – Displays the
identified direct targets for the gene
Regulatory Network – Displays the interaction
between the direct targets.
Links: you can view additional information from various
Arabidopsis resources TIGR,
MIPS, SALK, and TAIR.
For example, if a search was conducted
using MYB15, the following pages would be displayed utilizing the various link.
(M) for MIPS (Munich Institute for
Protein Sequences): the MATDB within the
MIPS website contains all Arabidopsis
sequences and annotations produced by the
Arabidopsis Genome Initiative plus
mitochondrial and chloroplast genomes.
Sequences were consolidated from European
sequencing projects, Cold Spring
Harbor Lab, and Washington University.
Information obtained from MIPS website includes name, title, function, PROSITE
motifs, and Arabidopsis ESTs (as seen in figure below).

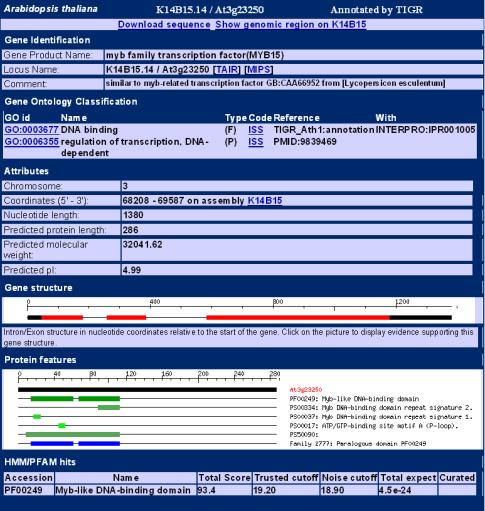
(T) for TIGR (The Institute for Genomic
Research): An alternative DB of
Arabidopsis sequences and annotations
from all AGI labs. The user can view various information about the gene
searched on, including Gene Product and Locus name, attributes, gene structure
and function (as seen in following figure).
(S) for SALK: Utilizing the SIGnAL
Arabidopsis Gene Mapping Tool, it provides a map of the genome area in which
the locus is found. On the map, cDNA (full-length cloned copies of mRNA) and
T-DNA (sites of insertion of Agrobacterium T-DNA in the Arabidopsis genome)
information is displayed in relation to the LocusID entered (as seen in figure
below).

(T) for TAIR (The Arabidopsis Information
Resource): A genomic data resource collected from a wide variety of sources,
with information about genes, markers, polymorphisms, maps, sequences, clones,
DNA and seed stocks, gene families and proteins. A link to the TAIR database
contains information on TAIR accession number, gene model, description,
nucleotide sequence, protein data, and map locations.